MUSHROOM BODY and control genes
Fruit flies exhibit a variety of cognitive behaviors including associative learning and memory. Consisting of 2500 neurons, mushroom body is a prominent brain structure in the fly brain and involved in a variety of neural functions. Anatomically, it receives olfactory information from the antennal lobe via olfactory projection neurons and thus may correspond to the olfactory cortex in the human brain. Molecular genetic studies have demonstrated that evolutionary conserved paired-type homeobox genes, eyeless/Pax6, play crucial roles in the differentiation of mushroom body neurons (Kurusu et al., 2000; Callaerts et al., 2001; Furukubo-Tokunaga et al., 2009). The vertebrate Pax6 gene is expressed in the olfactory cortex and amygdala, which are involved in olfactory perception and memory formation.
A genome-wide survey of the Drosophila mushroom body transcripts identified a large number of genes that are preferentially expressed in mushroom body neurons and thus may represent candidates of the downstream targets of the eyeless/Pax6 genes in brain development (Kobayashi et al., 2006). RNA interference analyses showed that many of the identified genes are required for the development of the MB lobes, while the rest of the genes are likely to be required for the plasticity and physiological functions of the mushroom body neurons (Kobayashi et al., 2006).
In addition, we have has demonstrated that an orphan nuclear receptor, Tailless (TLL), is required for prolonged cell division of the mushroom body neuroblast or stem cells (Kurusu et al., 2009). Intriguingly, the vertebrate homolog TLX (NR2E1) gene is expressed in the adult neurogenic regions including the subgranular layer of the dentate gyrus. These results suggest an intriguing commonality of the genetic programs that control the proliferation of neural stem cells of the brain structures involved in memory processing in both the Drosophila and the vertebrate brains.
Genetic program of mushroom body development
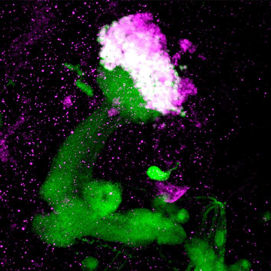
Mushroom body of the Drosophila brain
Green: mushroom body as revealed with a GAL4 driver (201Y) and GFP
Magenta: Eyeless/Pax6 protein expression in the nuclei of the mushroom body neurons
Confocal microscopy of an adult fly brain
Kurusu, M. et al., 2000.
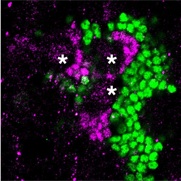
TLL controls proliferation of the mushroom body progenitor cells
Green: mushroom body neurons as revealed with an anti-DAC antibody staining
Magenta: TLL expression in the nuclei of the progenitor cells. Asterisks: neuroblasts.
Kurusu, M. et al., 2009.
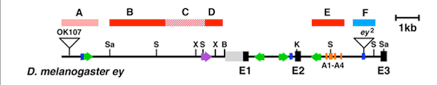
The cis-regulatory moduli of the Drosophila eyeless/Pax6 gene.
Eye-specific expression of the eyeless/Pax6 gene is driven by the intronic module (Box F) that harbors several paired-domain binding sites. On the other hand, the complex brain expression pattern of eyeless is driven by the coordination of multiple enhancer moduli (Box A-E).
Adachi, Y. et al., 2003.
Apart from conserved transcription factors, a variety of molecules participate in the formation of the elaborate mushroom body structures. Clonal studies showed that developing mushroom body has concentric layers, in which axons of newly born Kenyon cells firstly project into the core and shift to more distal layers as they undergo further differentiation (Kurusu et al., 2002). Regulatory proteins involved in growth cone guidance such as Fas II, an immunoglobulin family molecule, Dscam (Zhan et al., 2004) and receptor tyrosine phosphatases (Kurusu and Zinn, 2008) are preferentially expressed in the mushroom body core fibers. Moreover, Mochizuki et al. (2011) showed preferential expression of a conserved Ser/Thr kinase, UNC-51, and kinesin motor proteins in the core fibers, and suggested active intracellular transport along the elongating axons, which is essential for the establishment and maintenance of the polarity and compartmentalization of the mushroom body neurons.
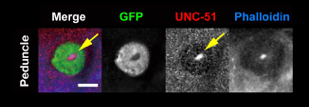
UNC-51 is localized to the core fibers in the developing mushroom body.
Green: 201Y GAL4
Red: anti-UNC-51 staining
Blue: actin fibers stained with phalloidin
Mochizuki et al., 2011
FURUKUBO-TOKUNAGA LABORATORY
INSTITUTE OF BIOLOGICAL SCIENCES, UNIVERSITY OF TSUKUBA, JAPAN
Update June 2018
